GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.
Python package for analysis of multiomic single cell RNA-seq and ATAC-seq. - GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.

MIRA: joint regulatory modeling of multimodal expression and chromatin accessibility in single cells

Integrated analysis of multimodal single-cell data - ScienceDirect
GitHub - databio/awesome-atac-analysis: Links to ATAC-seq analysis tools

MIRA: Joint regulatory modeling of multimodal expression and chromatin accessibility in single cells

A high-resolution CITE-seq atlas of the murine spleen. (A) UMAP

MultiMAP: dimensionality reduction and integration of multimodal data, Genome Biology
GitHub - mdozmorov/RNA-seq_notes: A continually expanding collection of RNA- seq tools
GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.

MIRA: Joint regulatory modeling of multimodal expression and chromatin accessibility in single cells

MIRA: Joint regulatory modeling of multimodal expression and chromatin accessibility in single cells
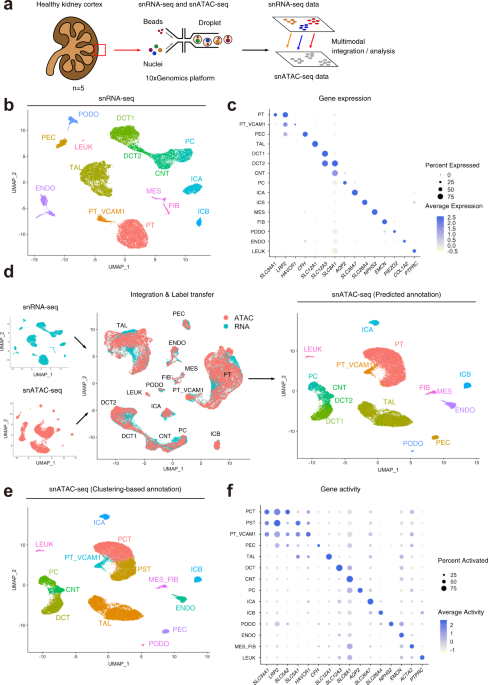
Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney
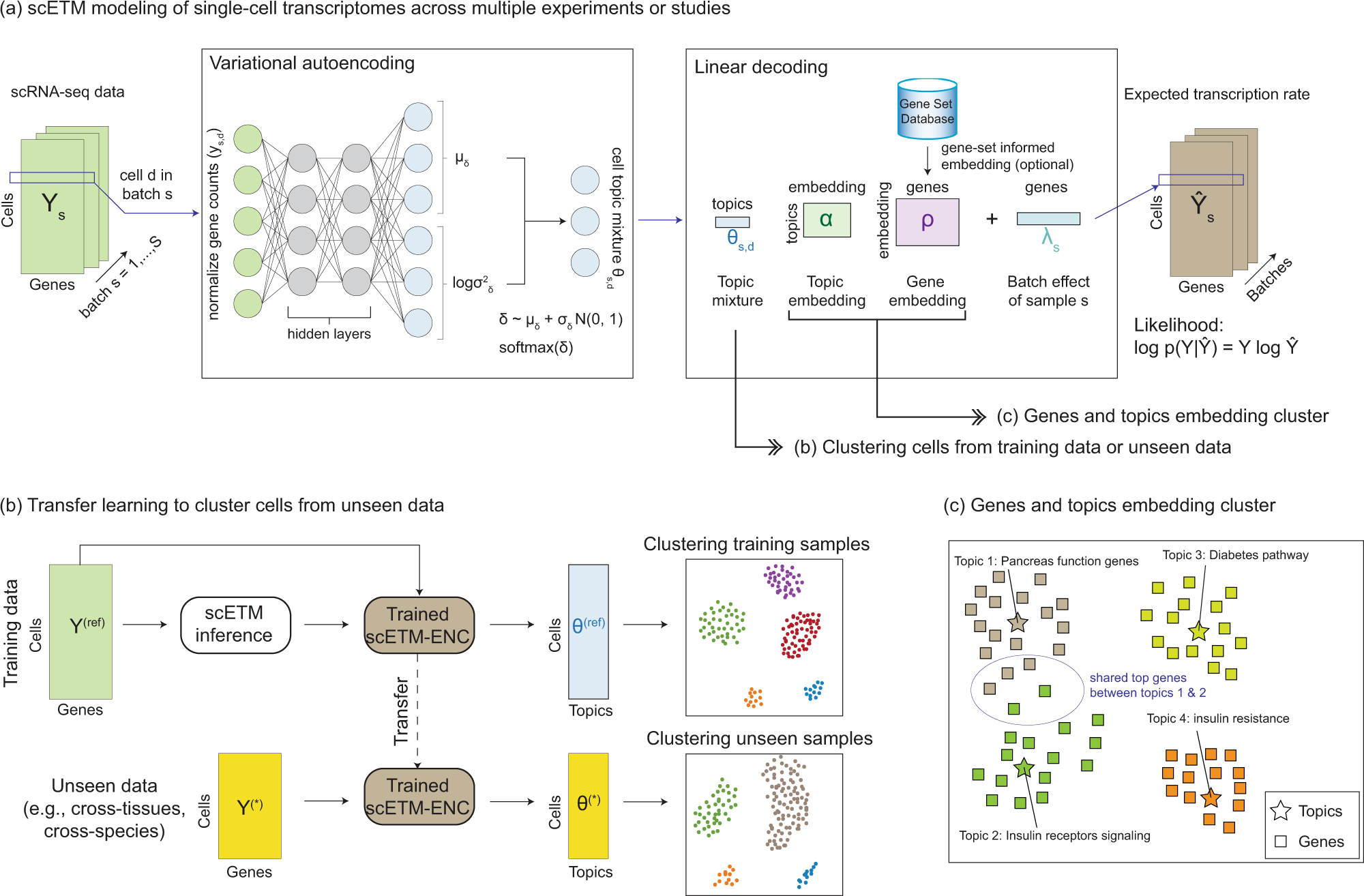
Learning interpretable cellular and gene signature embeddings from single- cell transcriptomic data
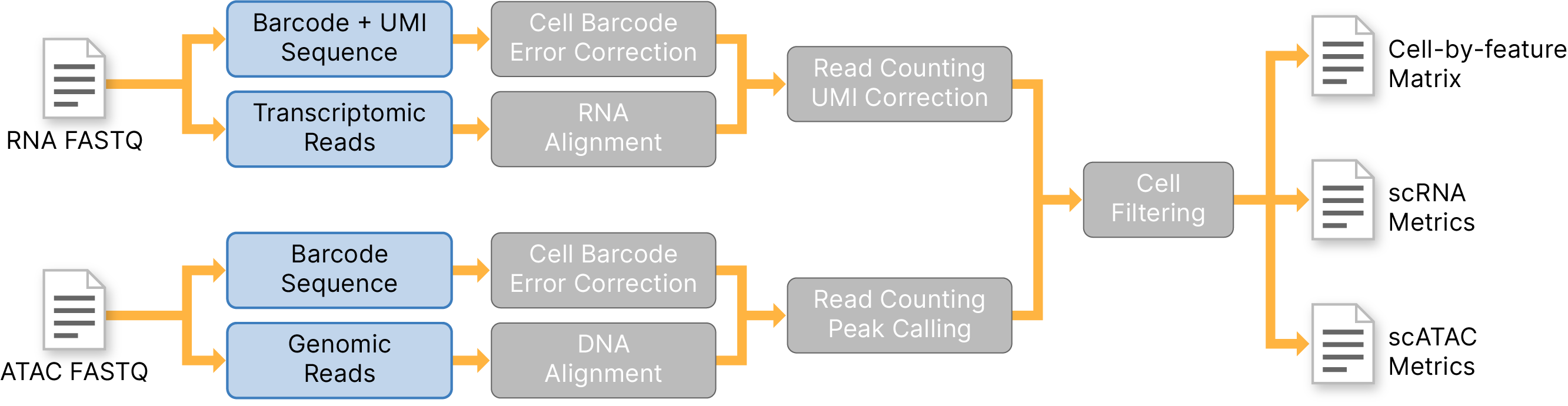
Multiomics Pipeline