
Expressions of the trajectory markers in bulk samples are
Download scientific diagram | Expressions of the trajectory markers in bulk samples are associatedwith poor prognosis in METABRIC and TCGA (A) Heatmap of top 500 tumor cluster marker enrichments in METABRIC TNBC patients. The main heatmap depicts GSVA enrichment score as sample-wise enrichments of the trajectory markers in METABRIC TNBC patients. On top of the heatmap, the annotations depict the number of lymph node metastasis, 5-year disease recurrence (relapse: red, no relapse: green) and TP53 mutation status. On the bottom, the abundance of stromal and immune cells by ESTIMATE score (low: yellow, high: red) is shown in the heatmap bar. The heatmap was clustered by complete linkage hierarchical clustering with 5 clusters as evaluated in Figure S4F. The poor prognosis associated with cluster 1 is highlighted to show enrichments of tumor cell markers from C3, C4, and C9. (B) Kaplan-Meier plot of METABRIC TNBC patients stratified by hierarchical clustering from the tumor cell marker enrichments in A. (C) Heatmap of marker GSVA z-scores in TCGA TNBC patients: From the top, the annotation depicts cancer stage, 5-year overall survival (death: red, alive: green), and sample clusters by hierarchical clustering on the GSVA matrix with k = 4. The ESTIMATE scores are shown on the bottom annotation bar. The tumor-enriched clusters with mild C9 and C2 marker enrichments are associated with poor prognosis and being highlighted in a red box. (D) Kaplan-Meier plot of TCGA TNBC patients stratified by hierarchical clustering (k = 4) from the marker enrichments in C. from publication: Pseudo-temporal dynamics of chemoresistant triple-negative breast cancer cells reveal EGFR/HER2 inhibition as synthetic lethal during mid-neoadjuvant chemotherapy | In the absence of targetable hormonal axes, chemoresistance for triple-negative breast cancer (TNBC) often compromises patient outcomes. To investigate the underlying tumor dynamics, we performed trajectory analysis on the single-nuclei RNA-seq (snRNA-seq) of chemoresistant | Chemoresistance, Synthetic Lethal Mutations and Triple Negative Breast Neoplasms | ResearchGate, the professional network for scientists.

Single-cell trajectory branch analysis of MSC subpopulations. a-c

Trajectory-based differential expression analysis for single-cell sequencing data

Single-cell transcriptomics identifies gene expression networks driving differentiation and tumorigenesis in the human fallopian tube - ScienceDirect

Single-cell gene expression analysis of human B cells a, t-SNE analysis
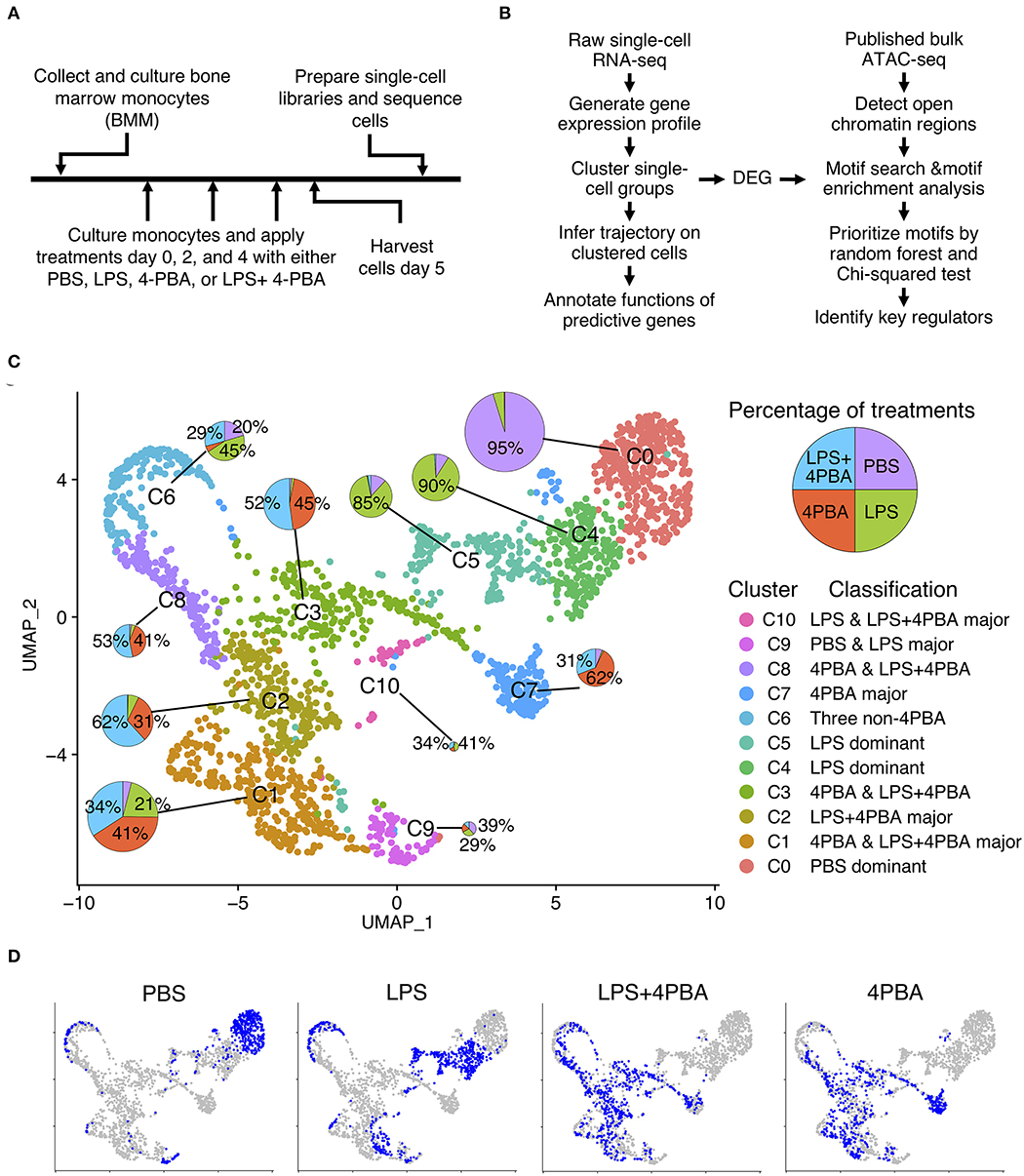
Frontiers Single Cell RNA-Seq and Machine Learning Reveal Novel Subpopulations in Low-Grade Inflammatory Monocytes With Unique Regulatory Circuits

Single-cell and spatial transcriptomics identify a macrophage population associated with skeletal muscle fibrosis

Targeted RNA expression profiling identifies high-grade endometrial stromal sarcoma as a clinically relevant molecular subtype of uterine sarcoma - Modern Pathology

PDF) Pseudo-temporal dynamics of chemoresistant triple-negative breast cancer cells reveal EGFR/HER2 inhibition as synthetic lethal during mid-neoadjuvant chemotherapy

Binary organization of epidermal basal domains highlights robustness to environmental exposure
Documentation - Gene Expression Nebulas

IJMS, Free Full-Text
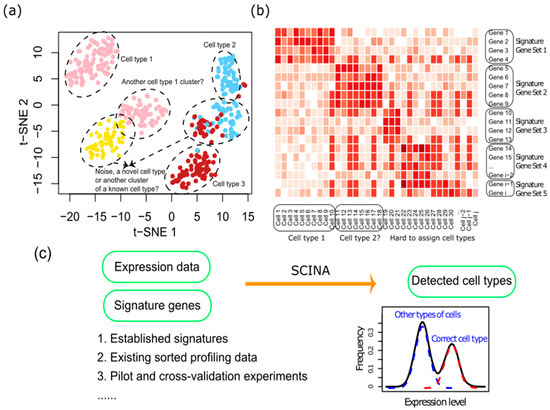
Genes, Free Full-Text








