
PICA: Pixel Intensity Correlation Analysis for Deconvolution and Metabolite Identification in Mass Spectrometry Imaging

Liron FELDBERG, PhD, Israel Institute of Biological Research, Ness Ziona, IIBR, Department of Analytical Chemistry

ISFrag: De Novo Recognition of In-Source Fragments for Liquid Chromatography–Mass Spectrometry Data

Metabolites, Free Full-Text

34676 PDFs Review articles in METABOLITE IDENTIFICATION

PICA: Pixel Intensity Correlation Analysis for Deconvolution and Metabolite Identification in Mass Spectrometry Imaging

Correlation-Based Deconvolution (CorrDec) To Generate High-Quality MS2 Spectra from Data-Independent Acquisition in Multisample Studies

Liron FELDBERG, PhD, Israel Institute of Biological Research, Ness Ziona, IIBR, Department of Analytical Chemistry
PDF) PICA: Pixel Intensity Correlation Analysis for Deconvolution and Metabolite Identification in Mass Spectrometry Imaging

rMSIannotation: A peak annotation tool for mass spectrometry imaging based on the analysis of isotopic intensity ratios - ScienceDirect

Enhanced in-Source Fragmentation Annotation Enables Novel Data Independent Acquisition and Autonomous METLIN Molecular Identification

PICA: Pixel Intensity Correlation Analysis for Deconvolution and Metabolite Identification in Mass Spectrometry Imaging

PICA: Pixel Intensity Correlation Analysis for Deconvolution and Metabolite Identification in Mass Spectrometry Imaging

Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights
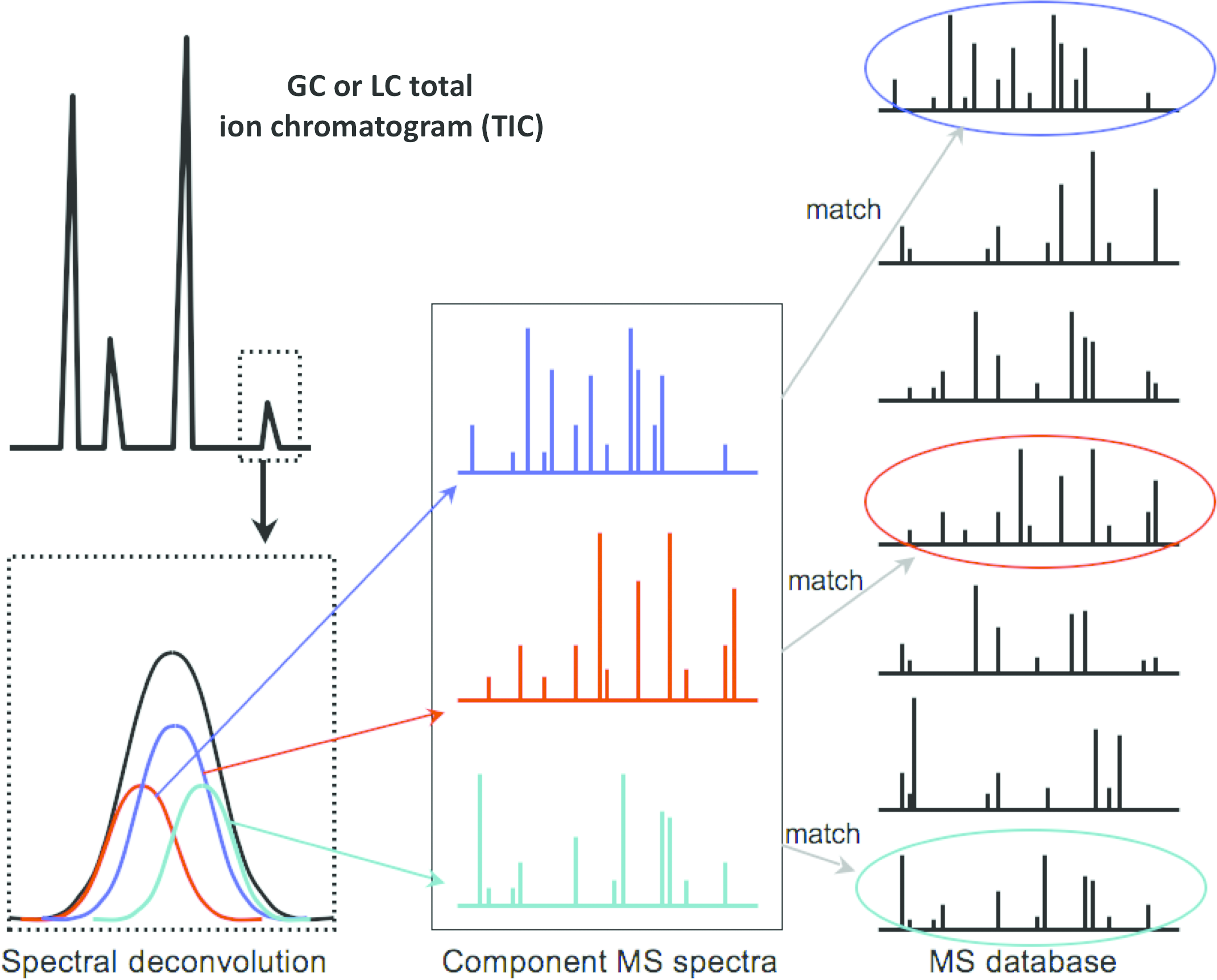
Exploring the Human Metabolome by Nuclear Magnetic Resonance Spectroscopy and Mass Spectrometry (Chapter 1) - Methodologies for Metabolomics








